 |
PING
0.9
Statistical data handling and processing in production environment
|
 |
PING
0.9
Statistical data handling and processing in production environment
|
The purpose of an adequate documentation is to enable the produser to (re)use, extend, share and migrate a program. It is also essential for business and processes modeling.
For that purpose, we adopt a standardised way for appropriately document PING programs and processes. In practice, an "inline" documentation:
and is then automatically extracted so as to create a user-friendly browsable "online" documentation using doxygen documentation generator.
We provide hereby guidelines and templates for actually documenting:
and we indicate as well how to automatically generate the documentation.
As mentioned above, the documentation of SAS programs is inserted in the header of the program as a comment. More precisely, we impose that:
/** and */.Further, we also require that:
/** \cond */ and /** \endcond */.You will also need to adopt a common template for documentation:
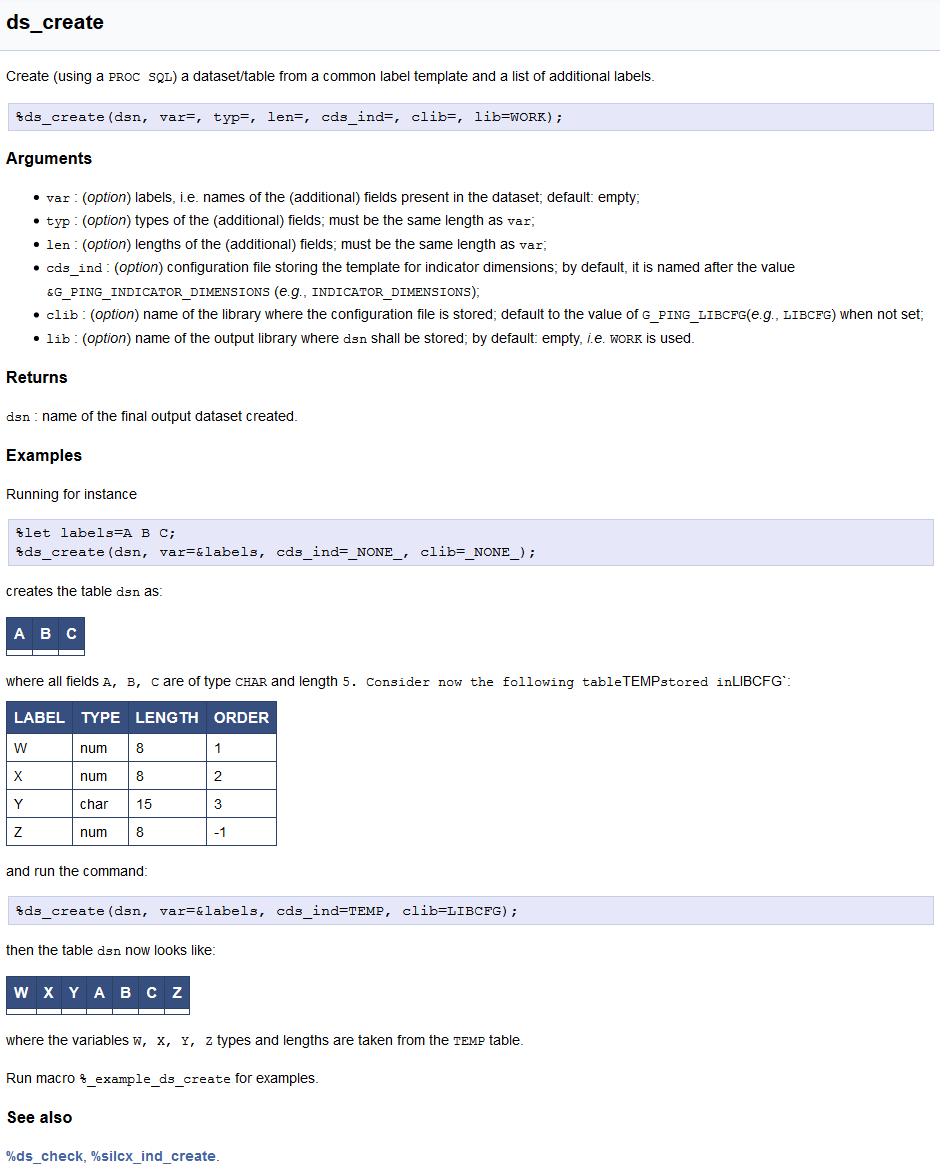
sas_### Arguments,### Returns,### Example (### Examples);sas shortname, hence ---sas or ~~~sas (both, in principle, supported for (pygmentation](http://pygments.org/docs/lexers/#lexer-for-sas),### Note (### Notes),### Reference (### References),PING or external) related programs/macros shall appear after the header ### See also.The inline documentation of the ds_create.sas macro looks like, for instance:
## ds_create {#sas_ds_create}
Create (using a `PROC SQL`) a dataset/table from a common label template and a list of
additional labels.
~~~sas
%ds_create(dsn, var=, typ=, len=, cds_ind=, clib=, lib=WORK);
~~~
### Arguments
* `var` : (_option_) labels, i.e. names of the (additional) fields present in the dataset;
default: empty;
* `typ` : (_option_) types of the (additional) fields; must be the same length as `var`;
* `len` : (_option_) lengths of the (additional) fields; must be the same length as `var`;
* `cds_ind` : (_option_) configuration file storing the template for indicator dimensions;
by default, it is named after the value `&G_PING_INDICATOR_DIMENSIONS` (_e.g._,
`INDICATOR_DIMENSIONS`);
* `clib` : (_option_) name of the library where the configuration file is stored; default
to the value of `G_PING_LIBCFG`(_e.g._, `LIBCFG`) when not set;
* `lib` : (_option_) name of the output library where `dsn` shall be stored; by default:
empty, _i.e._ `WORK` is used.
### Returns
`dsn` : name of the final output dataset created.
### Examples
Running for instance
~~~sas
%let labels=A B C;
%ds_create(dsn, var=&labels, cds_ind=_NONE_, clib=_NONE_);
~~~
creates the table `dsn` as:
| A | B | C |
|---|---|---|
| | | |
where all fields `A, B, C` are of type `CHAR` and length `5.
Consider now the following table `TEMP` stored in `LIBCFG`:
LABEL | TYPE | LENGTH | ORDER
------|------|--------|------
W | num | 8 | 1
X | num | 8 | 2
Y | char | 15 | 3
Z | num | 8 | -1
and run the command:
~~~sas
%ds_create(dsn, var=&labels, cds_ind=TEMP, clib=LIBCFG);
~~~
then the table `dsn` now looks like:
| W | X | Y | A | B | C | Z |
|---|---|---|---|---|---|---|
| | | | | | | |
where the variables `W, X, Y, Z` types and lengths are taken from the `TEMP` table.
Run macro `%%_example_ds_create` for examples.
### See also
[%ds_check](@ref sas_ds_check), [%silcx_ind_create](@ref sas_silcx_ind_create).
so that the online documentation is rendered as:
Similarly, the documentation shall be inserted in the header of the program as a comment, hence after the # symbol. In practice, you will further need to insert the desired documentation in-between two anchors: #STARTDOC and #ENDDOC so as to recognise the text as specific to the documentation (and differentiate from other comments).
The common template for code documentation is exactly the same as the one used for SAS, with the following exceptions:
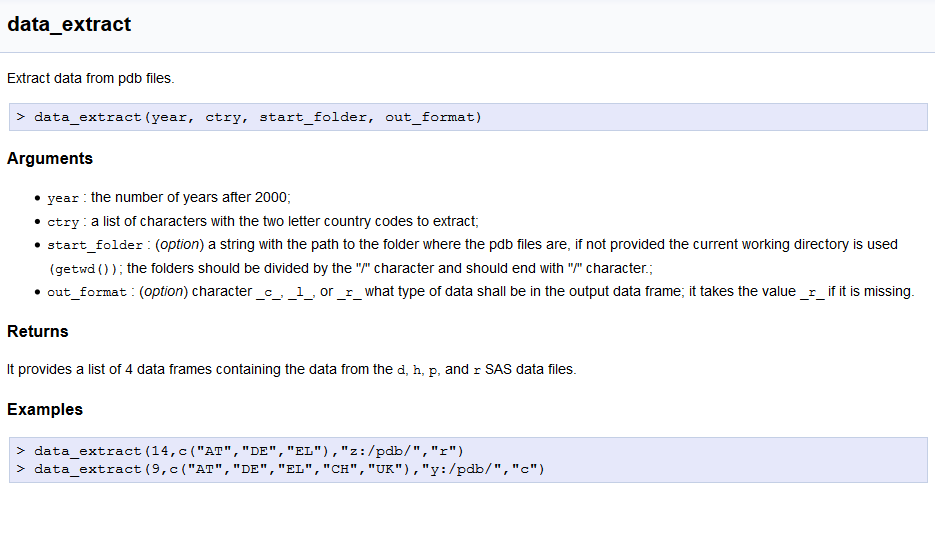
r_ is added (instead of sas_ above),> symbol (like in R console),r shortname, hence ---r or ~~~r (for (pygmentation](http://pygments.org/docs/lexers/#pygments.lexers.r.SLexer),As an example, the inline documentation inserted in the data_extract.R program looks like:
#STARTDOC
### data_extract {#r_data_extract}
#Extract data from pdb files.
#
#~~~r
# > data_extract(year, ctry, start_folder, out_format)
#~~~
#
#### Arguments
#* `year` : the number of years after 2000;
#* `ctry` : a list of characters with the two letter country codes to extract;
#* `start_folder` : (_option_) a string with the path to the folder where the pdb files are,
# if not provided the current working directory is used `(getwd())`; the folders should be
# divided by the "/" character and should end with "/" character.;
#* `out_format` : (_option_) character `_c_`, `_l_`, or `_r_` what type of data shall be in
# the output data frame; it takes the value `_r_` if it is missing.
#
#### Returns
# It provides a list of 4 data frames containing the data from the `d`, `h`, `p`, and `r`
# SAS data files.
#
#### Examples
#
#~~~r
# > data_extract(14,c("AT","DE","EL"),"z:/pdb/","r")
# > data_extract(9,c("AT","DE","EL","CH","UK"),"y:/pdb/","c")
#~~~
#ENDDOC
and the online documentation is rendered as:
So as to automatically generate the documentation (like this one), you will need:
markdown formatted documentation from program files into pure markdown files,markdown files.As for the documentation extractor, you can use a bash script (hereby named rsas2mddoc.sh) specifically developed for this purpose. This ad-hoc program enables you to retrieve automatically the markdown formatted documentation inserted in R/SAS files (as described above), and store the resulting excerpts into separated files.
This script is located under the documentation folder documentation\bin. It works as an inline command:
that can be launched from any terminal so as to generate a bulk of markdown files (with .md extension) into the documentation\md\library folder. The associated help looks like the following:
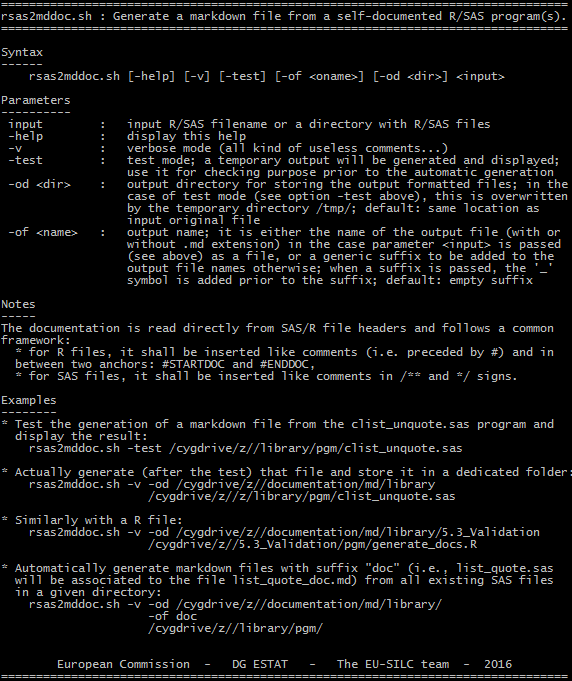
The resuiting markdown files will look exactly like the headers in your programs with the exception of the /* (or /**) and */ anchors. Then, doxygen is the tool used to actually generate the documentation. The full set of guidelines/best practices for running this software is available in the dedicated section of the doxygen website.