Usage
SAS programs
Compute the quartiles of a randomly generated vector (with normal distribution) using default parameters of the quantile function:
DATA test;
DO i = 1 TO 1000;
x = rand('NORMAL');
output;
END;
DROP i;
RUN;
%LET q_x=;
%quantile(x, idsn = test, _quantiles_ = q_x);
%PUT &q_x;
Do change the algorithm used for estimation:
%LET q_x=;
%quantile(x, type = 5, idsn = test, _quantiles_ = q_x);
%PUT &q_x;
Now compute the quintiles:
%LET q_x=;
%quantile(x, probs = 0.2 0.4 0.6 0.8, idsn = test, _quantiles_ = q_x);
%PUT &q_x;
Consider comparing the results obtained using both already existing PROC UNIVARIATE and the new implementation:
%LET probs = .2 .4 .6 .8;
%LET type = 8;
%quantile(x, probs=&probs, type=&type, method=DIRECT, _quantiles_ = q1);
%PUT &q1;
PROC UNIVARIATE DATA=test;
VAR x;
OUTPUT OUT=result PCTLPTS=&probs PCTLPRE=P_;
RUN;
DATA result;
SET result;
ARRAY P P_:;
DO i=1 TO 5;
CALL SYMPUT("q2","&q2 "!!LEFT(P(i)));
END;
RUN;
%PUT &q2;
Python programs
Compute the quartiles of a randomly generated vector (with normal distribution) using default parameters of the quantile function:
>>> import numpy as np
>>> x = np.random.rand(1000)
>>> from quantile import quantile
>>> q = quantile(x)
>>> print(q)
[0.10668975 0.19514584 0.33299627 0.53016722 0.89982259]
Do change the algorithm used for estimation:
>>> q = quantile(x, typ=5)
>>> print(q)
[0.10668975 0.19323867 0.33299627 0.54542127 0.89982259]
Now compute the quintiles:
>>> q = quantile(x, probs=[0., .2, .4, .6, .8, 1.])
>>> print(q)
[0.10668975 0.17672622 0.21760127 0.45610321 0.60035713 0.89982259]
Consider comparing the results obtained using both already existing scipy.mquantiles and the new implementation:
>>> probs = [0., .2, .4, .6, .8, 1.]
>>> typ = 8
>>> q1 = quantile(x, probs=probs, typ=typ, method='DIRECT')
>>> print(q1)
[0.10668975 0.14370132 0.21388262 0.46239251 0.71022886 0.89982259]
>>> q2 = quantile(x, probs=probs, typ=typ, method='INHERIT')
>>> print(q2)
[0.10668975 0.14370132 0.21388262 0.46239251 0.71022886 0.89982259]
It is possible to run (“call”) exactly the same estimations on an input file of sampled data, e.g.:
>>> ifile = "tests/sample1.csv"
>>> from io_quantile import IO_Quantile
>>> Q=IO_Quantile(probs=probs, typ=typ, method='DIRECT')
>>> q = Q(ifile)
>>> print(q)
[-3.71390789 -0.84013745 -0.27581729 0.1972615 0.73643897 2.84320792]
An instance of the class IO_Quantile is associated to one possible configuration of the quantile estimation. Usually, you will define another instance to perform the estimation with different parameters, e.g. using specialised quantiles:
>>> probs = "V20"
>>> Q=IO_Quantile(probs=probs, typ=7)
>>> q = Q(ifile)
>>> print(q)
[-1.77201688 -1.35332892 -1.02227673 -0.8369087 -0.65649296 -0.52217107
-0.40274654 -0.27525036 -0.17099489 -0.02049303 0.07102564 0.1971682
0.3350732 0.4536729 0.58860973 0.73606624 0.92265672 1.16084927
1.51058281]
But then it is possible to run the same estimation algorithm over different input files using that same, already defined, instance:
>>> ifile2 = "tests/sample2.csv"
>>> q2 = Q(ifile2)
>>> print(q2)
[-1.50530344 -1.1758764 -0.97947549 -0.82893958 -0.67830867 -0.52803119
-0.38669867 -0.27675766 -0.15487459 -0.01350255 0.13391096 0.27587611
0.40033506 0.54753533 0.68432219 0.84362724 1.05800707 1.34756166
1.73445305]
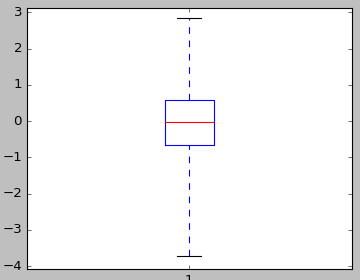
Note the definition of the IO_Quartile class that specifically runs estimation of quartiles, and also enables you to plot the associated boxplot:
>>> from matplotlib import pyplot
>>> from io_quantile import IO_Quartile
>>> Qu=IO_Quartile(typ=7);
>>> q = Qu(ifile)
>>> print(q)
[-3.71390789 -0.65649296 -0.02049303 0.58860973 2.84320792]
>>> Qu.plot(ifile)
>>> pyplot.show()
R programs
Compute the quartiles of a randomly generated vector (with normal distribution) using default parameters of the quantile function:
> source("quantile.R")
> x <- rnorm(1000)
> quantile(x)
[1] -3.4336152 -0.6638305 0.0228467 0.7090099 3.1428912
Note the usage of names parameter for formatting the output vector (likewise the original stats::quantile function):
> quantile(x, names=TRUE)
0% 25% 50% 75% 100%
-3.4336152 -0.6638305 0.0228467 0.7090099 3.1428912
Check that in INHERIT mode, we indeed wrap the original stats::quantile function:
> quantile(x, method='INHERIT')
0% 25% 50% 75% 100%
-3.4336152 -0.6638305 0.0228467 0.7090099 3.1428912
> stats::quantile(x)
0% 25% 50% 75% 100%
-3.4336152 -0.6638305 0.0228467 0.7090099 3.1428912
Do select other input parameters to test different types of implementation:
> quantile(x, type=5, probs=seq(0.,1.,0.1), names=TRUE)
0% 10% 20% 30% 40% 50% 60% 70% 80% 90% 100%
-3.4336152 -1.3146826 -0.8156684 -0.5113350 -0.2285216 0.0228467 0.2787606 0.5420263 0.8906395 1.3397724 3.1428912
You can for instance check the effect of the choice of the estimation algorithm on the calculation of the median:
> quantile(x, type=1, probs=0.5, names=TRUE)
50%
0.0198003
> quantile(x, type=3, probs=0.5, names=TRUE)
50%
0.0198003
> quantile(x, type=5, probs=0.5, names=TRUE)
50%
0.0228467
> stats::quantile(x, type=7, probs=0.5, names=TRUE)
50%
0.0228467
> quantile(x, type=7, probs=0.5, names=TRUE)
50%
0.0228467
> quantile(x, type=8, probs=0.5, names=TRUE)
50%
0.0228467
> quantile(x, type=10, probs=0.5, names=TRUE)
50%
0.0228467
Note that the method also works with data frame objects:
> d = data.frame(x=x)
> quantile(1, type=11, probs=0.5, names=TRUE, data=d)
50%
0.0228467
Finally, you can, likewise the Python implementation, run the quantile estimation on an input file
> source("io_quantile.R")
> ifile = "/Users/gjacopo/Developments/quantile/tests/sample1.csv"
> io_quantile(ifile, names=TRUE)
0% 25% 50% 75% 100%
-3.71390789 -0.65658941 -0.01801632 0.58892170 2.84320792